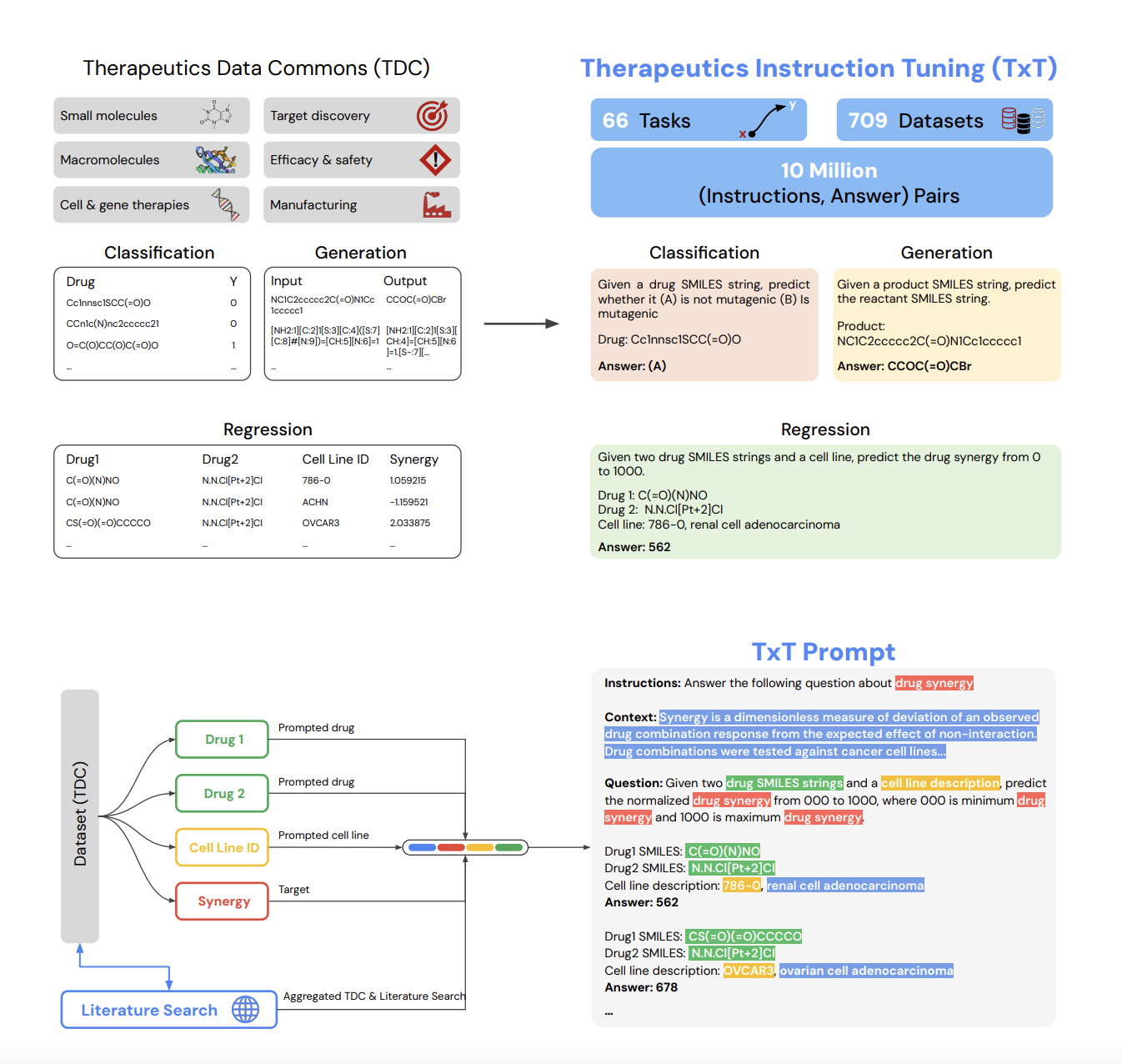
Developing therapeutics is costly and time-consuming, often taking 10-15 years and up to $2 billion, with most drug candidates failing during clinical trials. A successful therapeutic must meet various criteria, such as target interaction, non-toxicity, and suitable pharmacokinetics. Current AI models focus on specialized tasks within this pipeline, but their limited scope can hinder performance. The Therapeutics Data Commons (TDC) offers datasets to help AI models predict drug properties, yet these models work independently. LLMs, which excel at multi-tasking, provide the potential to improve therapeutic development by learning across diverse tasks using a unified approach.
LLMs, particularly transformer-based models, have advanced natural language processing, excelling in tasks through self-supervised learning on large datasets. Recent studies show LLMs can handle diverse tasks, including regression, using textual representations of parameters. In therapeutics, specialized models like graph neural networks (GNNs) represent molecules as graphs for functions such as drug discovery. Protein and nucleic acid sequences are also encoded to predict properties like binding and structure. LLMs are increasingly applied in biology and chemistry, with models like LlaSMol and protein-specific models achieving promising results in drug synthesis and protein engineering tasks.
Researchers from Google Research and Google DeepMind introduced Tx-LLM, a generalist large language model fine-tuned from PaLM-2, designed to handle diverse therapeutic tasks. Trained on 709 datasets covering 66 functions across the drug discovery pipeline, Tx-LLM uses a single set of weights to process various chemical and biological entities, such as small molecules, proteins, and nucleic acids. It achieves competitive performance on 43 tasks and surpasses state-of-the-art on 22. Tx-LLM excels in tasks combining molecular representations with text and shows positive transfer between different drug types. This model is a valuable tool for end-to-end drug development.
The researchers compiled a dataset collection called TxT, containing 709 drug discovery datasets from the TDC repository, focusing on 66 tasks. Each dataset was formatted for instruction tuning, featuring four components: instructions, context, question, and answer. These tasks included binary classification, regression, and generation tasks, with representations like SMILES strings for molecules and amino acid sequences for proteins. Tx-LLM was fine-tuned from PaLM-2 using this data. They evaluated the model’s performance using metrics such as AUROC and Spearman correlation and set accuracy. Statistical tests and data contamination analyses were performed to ensure robust results.
The Tx-LLM model demonstrated strong performance on TDC datasets, surpassing or matching state-of-the-art (SOTA) results on 43 out of 66 tasks. It outperformed SOTA on 22 datasets and achieved near-SOTA performance on 21 others. Notably, Tx-LLM excelled in datasets combining SMILES molecular strings with text features like disease or cell line descriptions, likely due to its pretrained knowledge of the text. However, it struggled on datasets that relied solely on SMILES strings, where graph-based models were more effective. Overall, the results highlight the strengths of fine-tuned language models for tasks involving drugs and text-based features.
Tx-LLM is the first LLM trained on diverse TDC datasets, including molecules, proteins, cells, and diseases. Interestingly, training with non-small molecule datasets, such as proteins, improved performance on small molecule tasks. While general LLMs have struggled with specialized chemistry tasks, Tx-LLM excelled in regression, outperforming state-of-the-art models in several cases. This model shows potential for end-to-end drug development, from gene identification to clinical trials. However, Tx-LLM is still in the research stage, with limitations in natural language instruction and prediction accuracy, requiring further improvement and validation for broader applications.
Check out the Paper and Details. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. If you like our work, you will love our newsletter.. Don’t Forget to join our 50k+ ML SubReddit
[Upcoming Event- Oct 17 202] RetrieveX – The GenAI Data Retrieval Conference (Promoted)
The post Google AI Introduces Tx-LLM: A Large Language Model (LLM) Fine-Tuned from PaLM-2 to Predict Properties of Many Entities that are Relevant to Therapeutic Development appeared first on MarkTechPost.