
Protein sequence design is crucial in protein engineering for drug discovery. Traditional methods like evolutionary strategies and Monte-Carlo simulations often need help to efficiently explore the vast combinatorial space of amino acid sequences and generalize to new sequences. Reinforcement learning offers a promising approach by learning mutation policies to generate novel sequences. Recent advancements in protein language models (PLMs), trained on extensive datasets of protein sequences, provide another avenue. These models score proteins based on biological metrics such as TM-score, aiding in protein design and folding predictions. These are essential for understanding cellular functions and accelerating drug development efforts.
Researchers from McGill University, Mila–Quebec AI Institute, ÉTS Montréal, BRAC University, Bangladesh University of Engineering and Technology, University of Calgary, CIFAR AI Chair, and Dreamfold propose using PLMs as reward functions for generating new protein sequences. However, PLMs can be computationally intensive due to their size. To address this, they introduce an alternative approach where optimization is based on scores from a smaller proxy model periodically fine-tuned alongside learning mutation policies. Their experiments across various sequence lengths demonstrate that RL-based approaches achieve favorable biological plausibility and sequence diversity results. They provide an open-source implementation facilitating the integration of different PLMs and exploration algorithms, aiming to advance research in protein sequence design.
Various methods have been explored for designing biological sequences. Evolutionary Algorithms like directed evolution and AdaLead focus on iteratively mutating sequences based on performance metrics. The Covariance Matrix Adaptation Evolution Strategy (CMA-ES) generates candidate sequences using a multivariate normal distribution. Proximal Exploration (PEX) promotes the selection of sequences close to wild type. Reinforcement Learning methods like DyNAPPO optimize surrogate reward functions to generate diverse sequences. GFlowNets sample compositions proportional to their reward functions, facilitating diverse terminal states. Generative Models like discrete diffusion and flow-based models like FoldFlow generate proteins in sequence or structure space. Bayesian Optimization adapts surrogate models to optimize sequences, addressing multi-objective protein design challenges. MCMC and Bayesian approach sample sequences based on energy models and structure predictions.
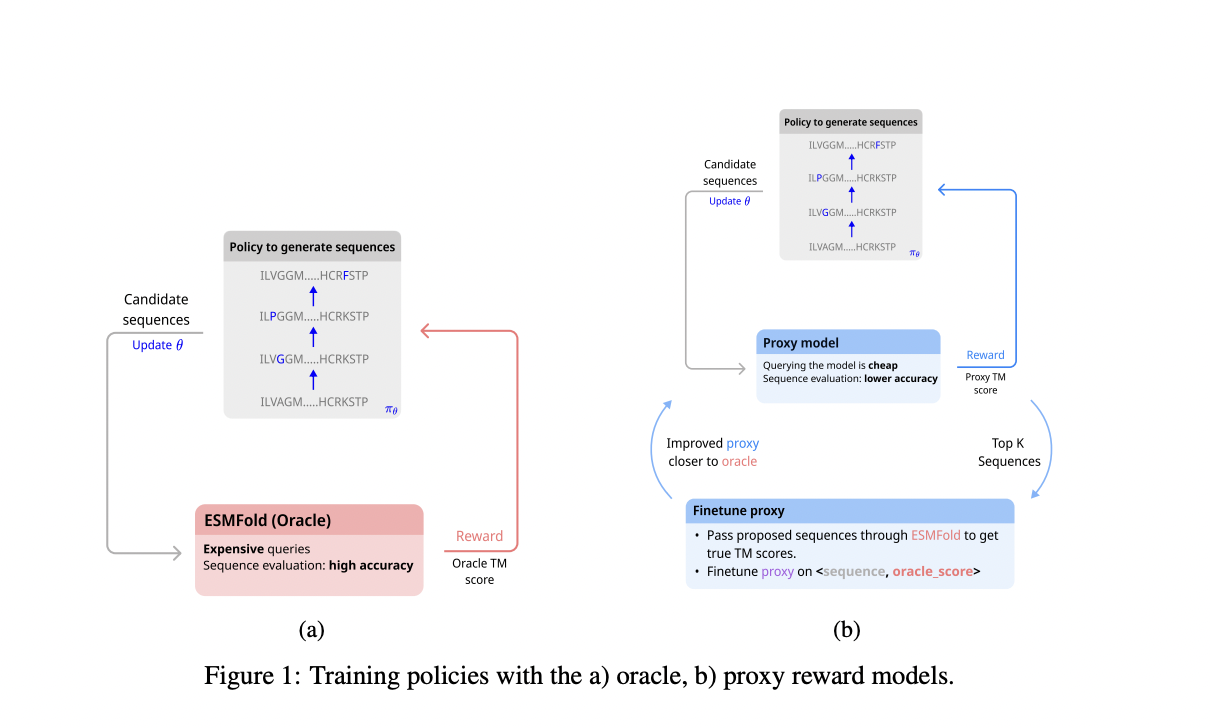
In the realm of protein sequence design using RL, the task is modeled as a Markov Decision Process (MDP) where sequences are mutated based on actions chosen by an RL policy. Sequences are represented in a one-hot encoded format, and mutations involve selecting positions and substituting amino acids. Rewards are determined by evaluating the structural similarity using either an expensive oracle model (ESMFold) or a cheaper proxy model periodically fine-tuned with true scores from the oracle. The evaluation criteria focus on biological plausibility and diversity, assessed through metrics like Template Modeling (TM) score and Local Distance Difference Test (LDDT), as well as sequence and structural diversity measures.
Various sequence design algorithms were evaluated using ESMFold’s pTM scores as the main metric in the experiments conducted. Results showed that methods such as MCMC excelled in directly optimizing pTM, while RL techniques and GFlowNets demonstrated efficiency by leveraging a proxy model. These methods maintained high pTM scores while significantly reducing computational costs. However, MCMC’s performance waned when finetuned with the proxy, possibly due to being trapped in suboptimal solutions aligned with the proxy model but not with ESMFold. Overall, RL methods like PPO and SAC, alongside GFlowNets, offered robust performance across bio-plausibility and diversity metrics, proving adaptable and efficient for sequence generation tasks.
The research findings are limited by computational constraints for longer sequences and reliance on either the proxy or the 3B ESMFold model for evaluation. Uncertainty or misalignment in the reward model adds complexity, necessitating future exploration with other PLMs like AlphaFold2 or larger ESMFold variants. Scaling to larger proxy models could enhance accuracy for longer sequences. While the study does not anticipate adverse implications, it highlights the potential misuse of PLMs. Overall, this work demonstrates the effectiveness of leveraging PLMs to develop mutation policies for protein sequence generation, showcasing deep RL algorithms as robust contenders in this field.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter.
Join our Telegram Channel and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 46k+ ML SubReddit
The post Advancements in Protein Sequence Design: Leveraging Reinforcement Learning and Language Models appeared first on MarkTechPost.