
Peptides, being highly flexible biomolecules, are involved in numerous biological processes and are of great interest in therapeutic development. Knowing the peptides’ conformations is crucial for any research as their function depends on their shape. Understanding how a peptide folds allows researchers to design new ones with specific therapeutic applications or helps them to deduce the processes by which natural peptides work at the molecular level, leading to advancements in various fields.
Researchers from the University of Toronto introduced PepFlow to address the challenge of accurately predicting the full range of conformations that peptides can assume. Traditional methods need help to effectively model the dynamic nature of peptides, enabling a more advanced approach to capture their various folding patterns and conformations.
Current methods for predicting biomolecular structures, like AlphaFold, have made significant advances in single-state prediction but fall short when dealing with the dynamic conformations of peptides. AlphaFold2, for instance, excels at predicting static protein structures but is not designed to generate a range of peptide conformations. This limits the understanding and utilization of peptides in biological and therapeutic contexts.
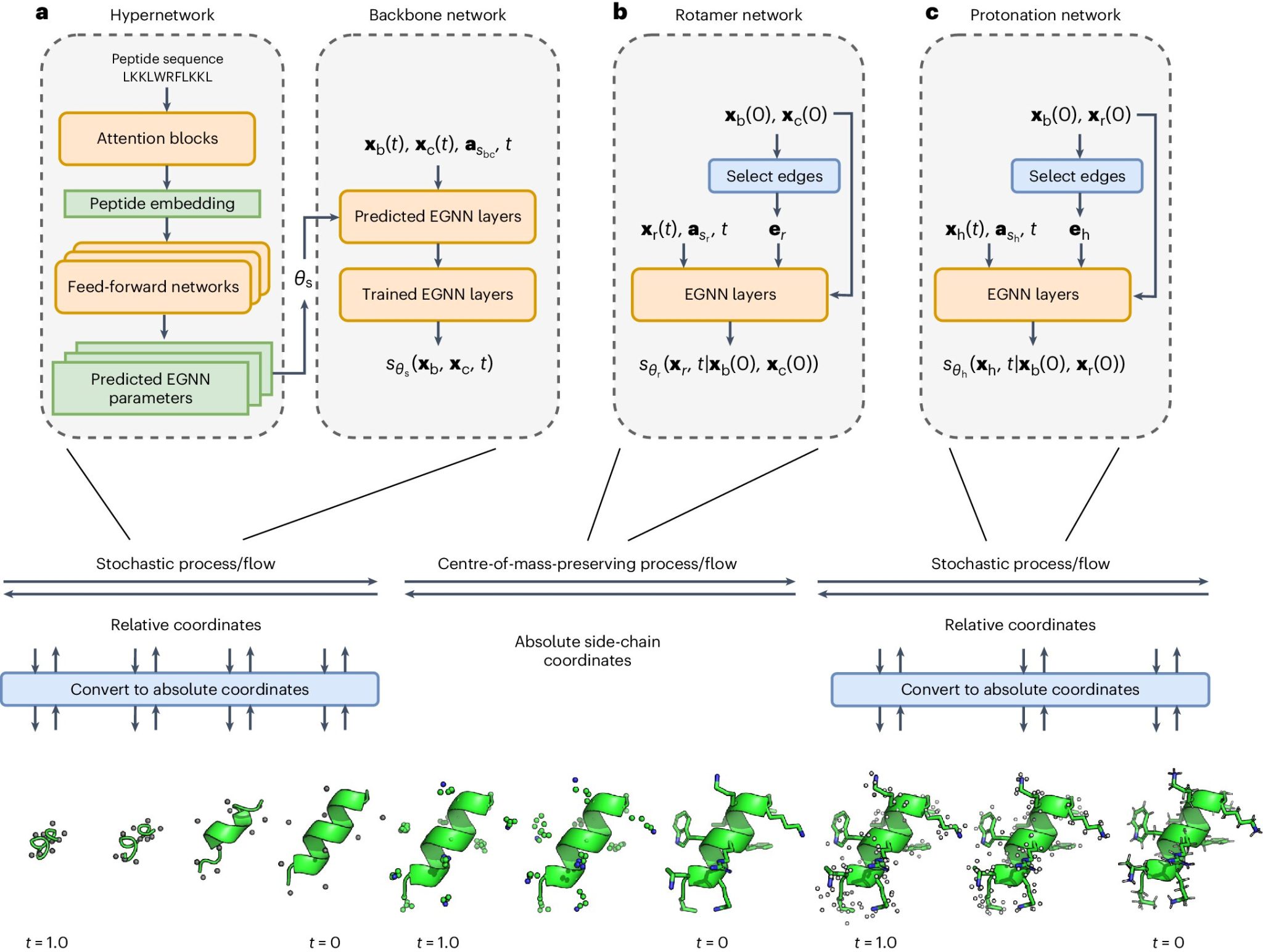
PepFlow is a deep-learning model explicitly designed to predict the full range of peptide conformations. PepFlow leverages a diffusion framework and integrates a hypernetwork to predict sequence-specific network parameters, enabling it to perform direct all-atom sampling from the allowable conformational space of peptides. This approach allows PepFlow to model peptide structures accurately and efficiently, surpassing the capabilities of current methods like AlphaFold2.
PepFlow combines machine learning with physics-based modeling to capture the dynamic energy landscape of peptides. The model is trained in a diffusion framework, which involves gradually transforming a simple initial distribution into a complex target distribution through a series of learned steps. This process allows PepFlow to generate diverse peptide conformations efficiently. A hypernetwork is employed to predict sequence-specific parameters, ensuring the model’s capability to adapt to different peptide sequences and their unique folding patterns.
One of the key innovations of PepFlow is its modular approach to generation, which helps mitigate the prohibitive computational cost associated with generalized all-atom modeling. By breaking down the generation process and using a hypernetwork, PepFlow can achieve high accuracy and efficiency. The model can predict peptide structures and recapitulate experimental peptide ensembles at a fraction of the running time required by traditional methods.
PepFlow’s performance is notable for its ability to model unusual peptide formations, such as macrocyclization, where peptides form ring-like structures. Such capabilities are valuable for drug development, as peptide macrocycles are a promising area of research for therapeutic applications. PepFlow demonstrates significant improvements over existing models, offering a comprehensive and efficient solution for peptide conformational sampling.
In conclusion, PepFlow addresses the challenge of predicting the full range of peptide conformations. By combining deep learning with physics-based modeling, PepFlow offers a highly accurate and efficient method for capturing the dynamic nature of peptides. This innovation not only surpasses current methods like AlphaFold2 but also holds significant potential for advancing therapeutic development through the design of peptide-based drugs. The study contains areas for further improvement, such as training with explicit solvent data, but PepFlow’s current capabilities mark a substantial advancement in biomolecular modeling.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter.
Join our Telegram Channel and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 45k+ ML SubReddit
The post Researchers at the University of Toronto Introduce a Deep-Learning Model that Outperforms Google AI System to Predict Peptide Structures appeared first on MarkTechPost.