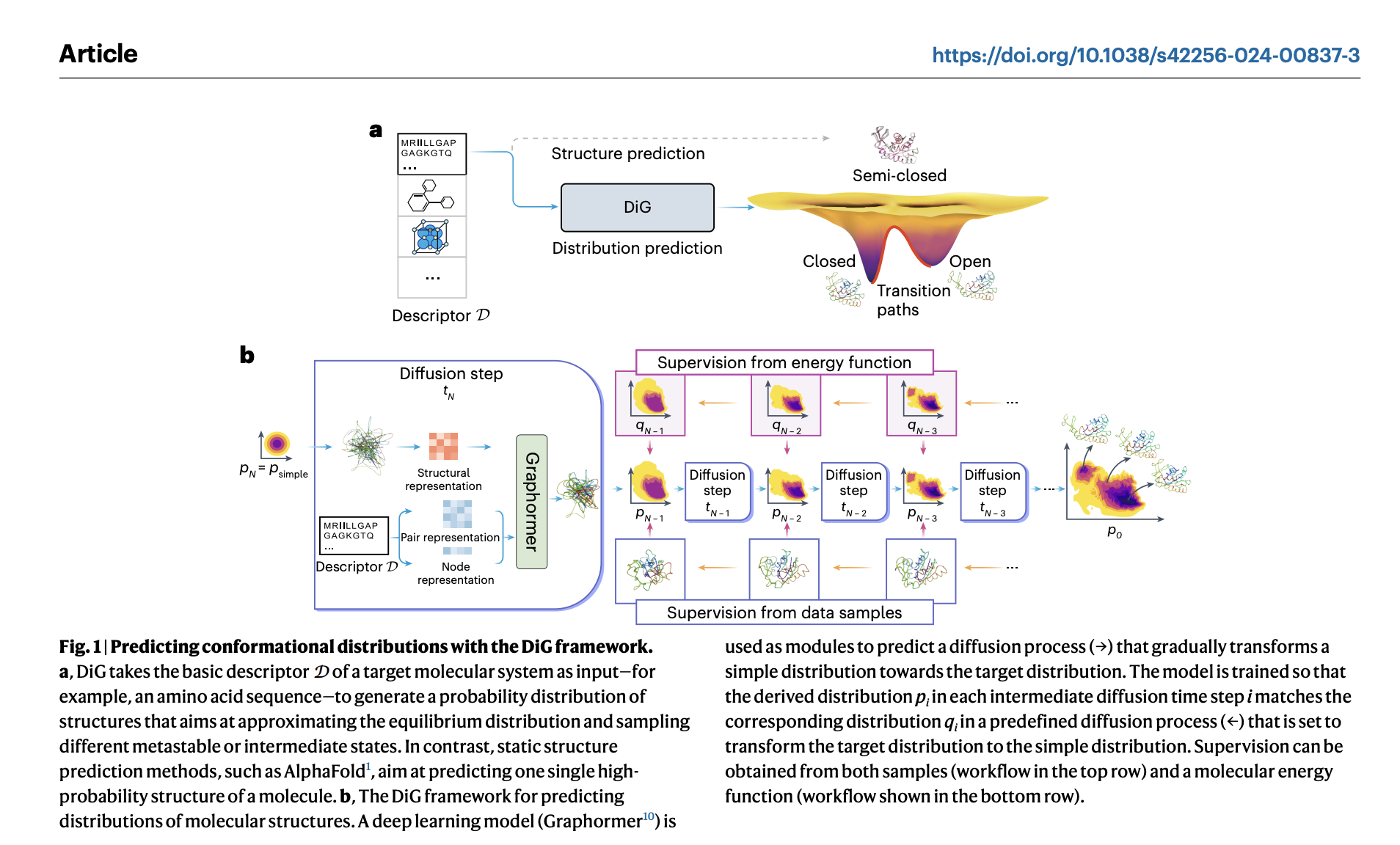
Advances in deep learning have revolutionized molecule structure prediction, but real-world applications often require understanding equilibrium distributions rather than just single structures. Current methods, like molecular dynamics simulations, are computationally intensive and insufficient for capturing the full range of molecular flexibility. Equilibrium distribution prediction is crucial for assessing macroscopic properties and functional states of molecules like adenylate kinase. While deep learning has shown promise in coarse-grained simulations, it struggles with generalization. Boltzmann generators offer a potential solution by generating equilibrium distributions, but their applicability across different molecules still needs to be improved.
Researchers from Microsoft Research AI4Science, Beijing, China; University of Science and Technology of China, Microsoft Quantum, Redmond, WA, USA; and Microsoft Research AI4Science, Berlin, Germany, have developed Distributional Graphormer (DiG), a deep learning framework aimed at predicting the equilibrium distribution of molecular systems. Inspired by thermodynamic annealing, DiG employs neural networks to transform a simple distribution towards equilibrium based on molecular descriptors like chemical graphs or protein sequences. This enables efficient generation of diverse conformations and estimations of state densities significantly faster than traditional methods. DiG demonstrates versatility across various molecular tasks and can generalize across different molecular systems. DiG approximates the equilibrium distribution by simulating a diffusion process, facilitating the prediction of molecular properties and enabling the inverse design of structures with desired properties.
DiG, a deep learning framework, extends beyond predicting single molecular structures to estimating their equilibrium distributions. Inspired by the heating-annealing concept, it employs a diffusion process to transform the target distribution towards a simpler one and then reverses it. Deep neural networks predict the reverse process by approximating the score function, facilitating the generation of diverse molecular structures. DiG also enables property-guided structure generation and interpolation between states by mapping structures to a latent space. This innovative approach advances molecular structure modeling, offering efficient predictions of equilibrium distributions and facilitating property-guided design.
DiG showcases its versatility by successfully tackling various molecular modeling and design challenges. For protein conformation sampling, it adeptly generates diverse structures consistent with the energy landscape, which is crucial for understanding protein behaviors and interactions. By leveraging experimental and simulated data, along with innovative training methods like PIDP, DiG accurately reproduces complex conformational distributions, even for proteins with multiple functional states. Furthermore, it demonstrates its ability to interpolate between states, providing insight into conformational transition pathways.
Expanding its scope, DiG excels in ligand structure sampling around binding sites, accurately predicting ligand structures within druggable pockets. Its performance, validated against experimental data, underscores its potential for drug design applications. Additionally, DiG proves its mettle in catalyst-adsorbate sampling, efficiently identifying active adsorption sites on catalyst surfaces. Its predictions align closely with those obtained through computationally intensive methods like density functional theory, highlighting its speed and accuracy. Lastly, DiG showcases its capability for property-guided structure generation, enabling inverse design tasks such as carbon allotrope generation with desired electronic band gaps. This demonstrates its potential to accelerate materials discovery and design processes.
In conclusion, DiG revolutionizes molecular sciences by predicting equilibrium distributions efficiently, enabling diverse molecular sampling crucial for understanding structure-function relationships and designing molecules and materials. DiG learns molecular representations from descriptors like protein sequences or compound formulas by employing advanced deep learning architectures, accurately capturing complex distributions in high-dimensional space. Its speed advantage over traditional methods like MD simulations or MCMC sampling offers transformative potential, reducing computational costs significantly. With its capacity to explore vast conformational spaces, DiG accelerates the discovery of molecular structures, impacting diverse fields, including life sciences, drug design, catalysis, and materials science.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter. Join our Telegram Channel, Discord Channel, and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 42k+ ML SubReddit
The post Microsoft Researchers Propose DiG: Transforming Molecular Modeling with Deep Learning for Equilibrium Distribution Prediction appeared first on MarkTechPost.