Medical LLMs like ClinicalCamel 70B and Llama3-OpenBioLLM 70B have shown strong performance in various medical NLP tasks, but no model specifically tailored to the cancer domain currently exists. Additionally, these models, with billions of parameters, are computationally demanding for many healthcare systems. A cancer-focused LLM, integrating specialized cancer knowledge, could significantly improve diagnosis and treatment planning. However, the high computational requirements of existing models highlight the need for smaller, more efficient LLMs accessible to healthcare institutions with limited resources, enabling broader adoption and better patient care in cancer treatment.
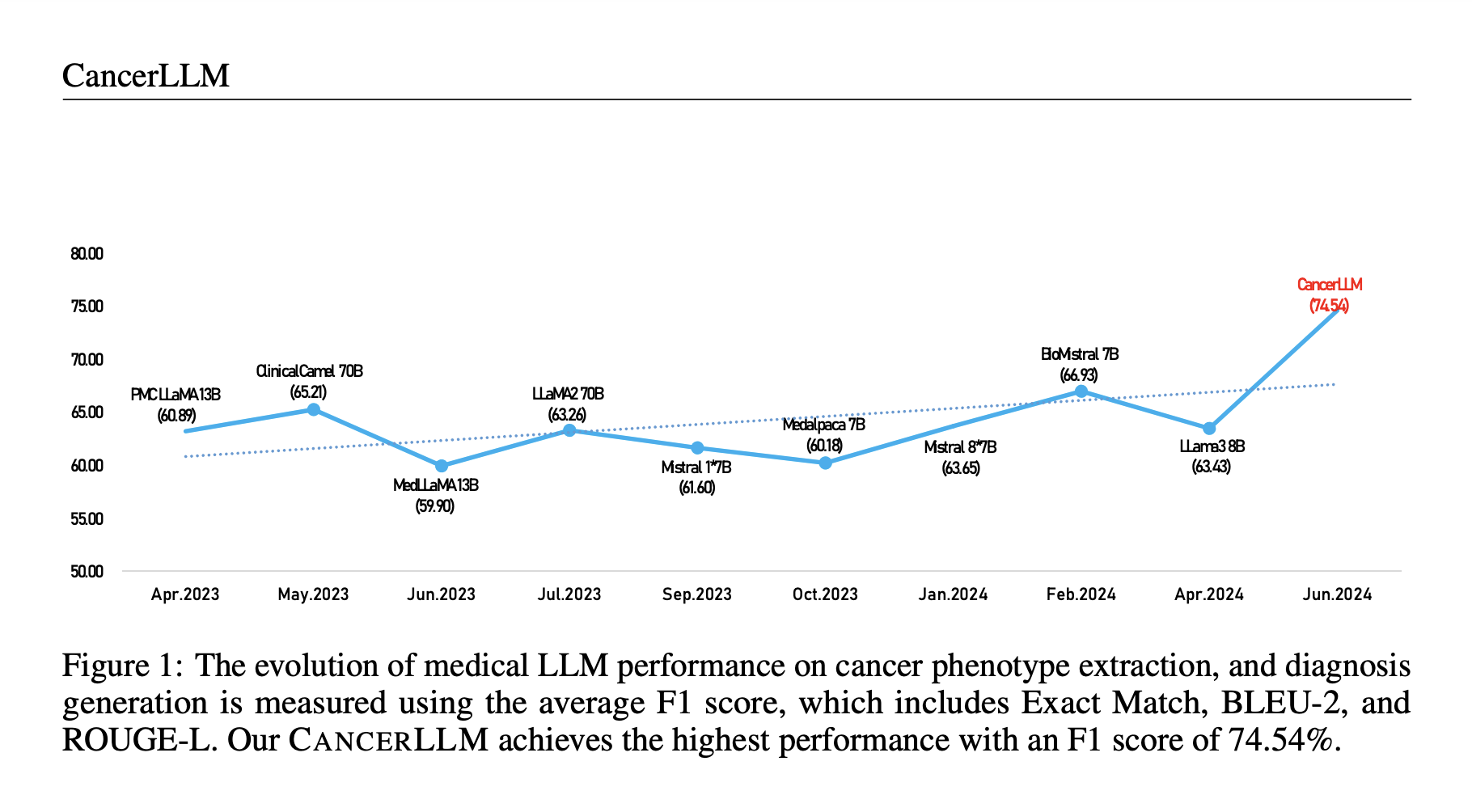
Researchers from multiple institutions, including the University of Minnesota and Yale, introduced CancerLLM, a 7-billion-parameter language model based on the Mistral architecture. Pre-trained on over 2.6 million clinical notes and 500,000 pathology reports covering 17 cancer types, CancerLLM was fine-tuned for cancer phenotype extraction and diagnosis generation tasks. It outperformed existing models by 7.61% on F1 scores and excelled in robustness tests involving counterfactuals and misspellings. This model aims to improve cancer research and healthcare delivery by enhancing clinical AI systems, providing a benchmark for cancer-focused tasks, and offering a robust, efficient tool for medical professionals.
CancerLLM’s workflow starts with injecting cancer-specific knowledge, followed by instruction tuning. Using clinical notes and pathology reports from 31,465 patients, the model was pre-trained to extract cancer phenotypes, generate diagnoses, and propose treatment plans. CancerLLM identifies entities such as tumor size, type, and stage for phenotype extraction. Diagnosis generation uses clinical notes to predict cancer diagnoses. The model was fine-tuned and evaluated using metrics like Exact Match, BLEU-2, and ROUGE-L, outperforming 14 baseline models and demonstrating robustness to counterfactuals and misspellings, achieving significantly higher precision, recall, and F1 scores compared to existing medical LLMs.
The results demonstrate that CANCERLLM significantly outperforms other medical LLMs across multiple tasks, including cancer diagnosis generation and phenotype extraction. It improves over baselines like Mistral 1*7B and Bio-Mistral 7B, with a notable 28.93% and 17.92% improvement in F1 scores for diagnosis generation, respectively. CANCERLLM also achieves better results than LLMs with larger parameters, like Llama3-OpenBioLLM-70B and ClinicalCamel-70B, indicating that model size alone is insufficient without domain-specific knowledge. Additionally, it proves robust against counterfactual errors and misspellings, maintaining performance across various rates of input errors. Its favorable balance of GPU memory usage and generation time highlights the model’s efficiency. It outperforms larger models like ClinicalCamel-70B regarding resource consumption while delivering competitive or superior results.
In the cancer diagnosis generation task, CancerLLM was pre-trained and fine-tuned to assess its effectiveness. Despite Bio-Mistral 7B achieving the best performance among baseline models, largely due to its extensive pre-processing on medical corpora like PubMed Central, CancerLLM outperformed all models, including the MoE-based Mistral 8*7B. Notably, even models with larger parameters, such as Llama3-OpenBioLLM-70B and ClinicalCamel-70B, did not surpass Bio-Mistral 7B, underscoring that more than larger parameter sizes are needed to guarantee better outcomes. The results reveal that CancerLLM’s domain-specific knowledge contributed to its superior performance, especially in cancer diagnosis generation. However, the model’s accuracy dropped when tasked with more complex diagnoses like ICD-based diagnosis generation. Despite this, CancerLLM demonstrated efficiency by reducing training time and resource usage, offering significant benefits to medical institutions with limited computational resources.
In the cancer phenotype extraction task, while ClinicalCamel-70B showed the best F1 score, its large size resulted in slower training and inference times. CancerLLM, with its smaller parameter size, performed comparably to Mistral 8*7B and Llama2 13B, reflecting its efficiency in resource-constrained environments. The model’s effectiveness was attributed to using clinical notes and pathology reports during training, though limited annotation data may have hindered performance in phenotype extraction. CancerLLM also demonstrated robustness against counterfactual errors and misspellings, though its performance decreased when misspelling rates exceeded 60%. These findings highlight the importance of high-quality annotation, meticulous data preprocessing, and the need to address specific challenges, such as misspellings and contextual misinterpretation, to improve CancerLLM’s diagnostic capabilities further.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and LinkedIn. Join our Telegram Channel.
If you like our work, you will love our newsletter..
Don’t Forget to join our 50k+ ML SubReddit
The post CancerLLM: A Large Language Model in Cancer Domain appeared first on MarkTechPost.